Overview
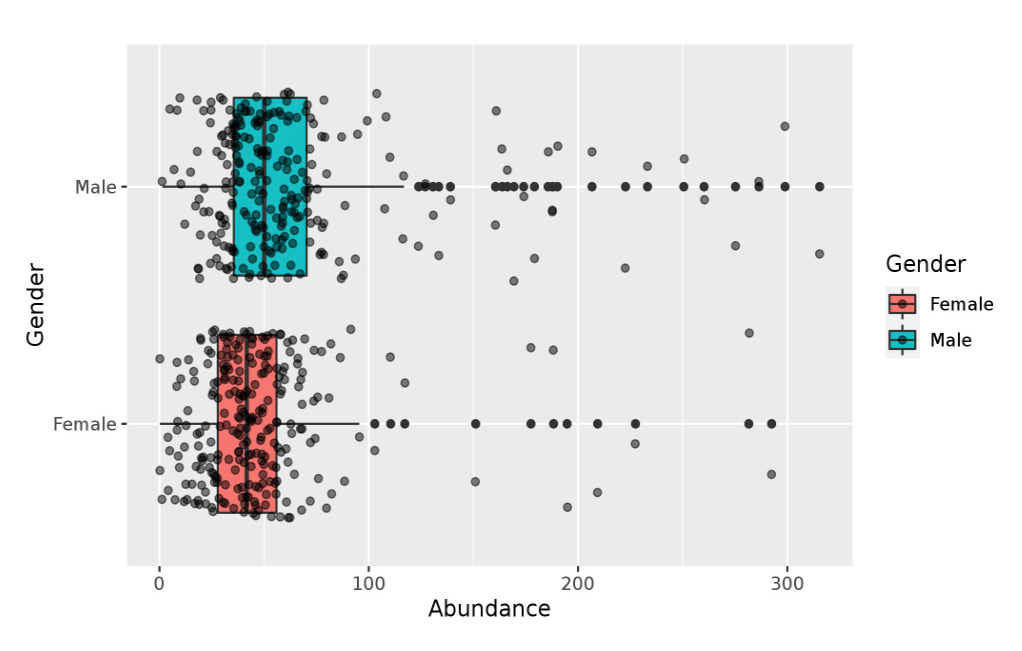
sBGC-hm is a database of biosynthetic gene clusters (BGCs) associated with the production of secondary metabolites in the human gut microbiome. These BGCs are involved in the production of a wide variety of molecules, including antibiotics and other small molecules that can have important biological functions. The database currently contains information on over 36,000 BGCs, with the most common type being ranthipeptides. The website provides several ways for users to access the information, including keyword searches, browsing, and BLAST searches. Each BGC is annotated with information such as the family it belongs to, the abundance of its genes, gender differences, transporter genes, and resistance genes. This resource provides valuable information for researchers studying the functions of the human gut microbiome and its potential applications in medicine.
Search
The quick search on the home page allows you to search individual fields found with secondary metabolite BGCs:
- Find a list of all indexed fields in the drop down menu and choose one of your interested.
- Enter the appropriate contents in the text area below.
- Click "Submit".

The search page allows you to find secondary metabolite BGCs with more than one fields by the logical operators "AND", "OR" and "NOT".
- Enter or select the items of your interested for your search.
- choose the appropriate logical operators behind.
- Click "Submit" (or click "Reset" to clear your input).
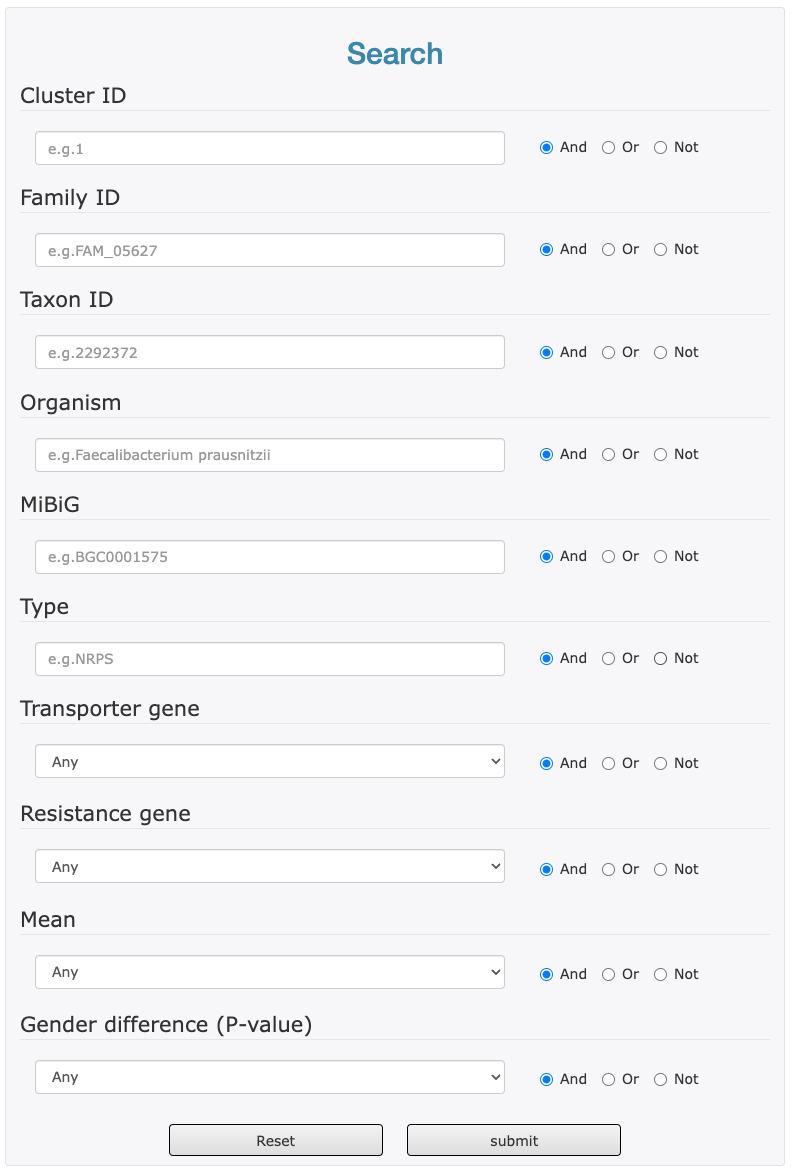
Search terms:
Type: BGC Type. The following table shows the type information included in current database. (For more information see https://docs.antismash.secondarymetabolites.org/glossary). e.g. NRPS or PKS
| Lable | Description |
|---|---|
| acyl_amino_acids | N-acyl amino acid |
| amglyccycl | Aminoglycoside/aminocyclitol |
| arylpolyene | Aryl polyene |
| betalactone | Beta-lactone containing protease inhibitor |
| butyrolactone | Butyrolactone |
| CDPS | tRNA-dependent cyclodipeptide synthases |
| cyclic-lactone-autoinducer | agrD-like cyclic lactone autoinducer peptides (AF001782) |
| ectoine | Ectoine |
| epipeptide | D-amino-acid containing RiPPs such as yydF (D78193) |
| furan | Furan |
| glycocin | Glycocin |
| hglE-KS | Heterocyst glycolipid synthase-like PKS |
| hserlactone | Homoserine lactone |
| ladderane | Ladderane |
| lanthipeptide-class-i | Class I lanthipeptides like nisin |
| lanthipeptide-class-ii | Class II lanthipeptides like mutacin II (U40620) |
| lanthipeptide-class-iii | Class III lanthipeptides like labyrinthopeptin (FN178622) |
| lanthipeptide-class-iv | Class IV lanthipeptides like venezuelin (HQ328852) |
| lanthipeptide-class-v | Glycosylated lanthipeptide/linaridin hybrids like MT210103 |
| LAP | Linear azol(in)e-containing peptides |
| lassopeptide | Lasso peptide |
| linaridin | Linear arid peptide such as cypemycin (HQ148718) and salinipeptin (MG788286) |
| microviridin | Microviridin |
| NAGGN | N-acetylglutaminylglutamine amide |
| NAPAA | Non-alpha poly-amino acids like e-Polylysin |
| NRPS | Non-ribosomal peptide synthetase |
| NRPS-like | NRPS-like fragment |
| nucleoside | Nucleoside |
| other | Cluster containing a secondary metabolite-related protein that does not fit into any other category |
| phenazine | Phenazine |
| phosphonate | Phosphonate |
| PKS-like | Other types of PKS |
| prodigiosin | Serratia-type non-traditional PKS prodigiosin biosynthesis pathway |
| proteusin | Proteusin |
| ranthipeptide | Cys-rich peptides (aka. SCIFF: six Cys in fourty-five) like in CP001581:3481278-3502939 |
| RaS-RiPP | Streptide-like thioether-bond RiPPs |
| redox-cofactor | Redox-cofactors such as PQQ (NC_021985:1458906-1494876) |
| resorcinol | Resorcinol |
| RiPP-like | Other unspecified ribosomally synthesised and post-translationally modified peptide product (RiPP) |
| RRE-containing | RRE-element containing cluster |
| sactipeptide | Sactipeptide |
| siderophore | Siderophore |
| T1PKS | Type I PKS (Polyketide synthase) |
| T2PKS | Type II PKS |
| T3PKS | Type III PKS |
| terpene | Terpene |
| thioamide-NRP | Thioamide-containing non-ribosomal peptide |
| thioamitides | Thioamitide RiPPs as found in JOBF01000011 |
| thiopeptide | Thiopeptide |
| transAT-PKS | Trans-AT PKS |
| transAT-PKS-like | Trans-AT PKS fragment, with trans-AT domain not found |
| tropodithietic-acid | Tropodithietic acid |
Blast
The BLAST (Basic Local Alignment Search Tool) program uses a strategy based on matching sequence fragments by employing a powerful statistical model to find the best local alignments (For more information see http://www.ebi.ac.uk/Tools/sss/ncbiblast/).
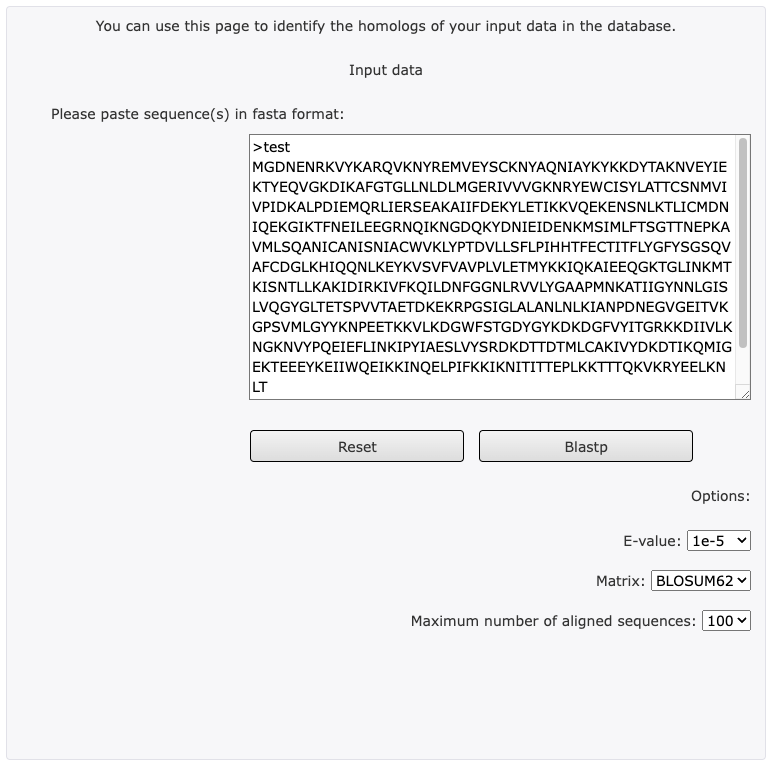
Step 1 – Sequence Input
- Sequence Input Window: The query sequence can be entered directly into text area. The sequence must be FASTA format.
FASTA format: FASTA formatted sequence records start with a definition line, which must start with a > character. The definition line must occupy one single line and followed by sequence data.
Example:
>test
MGDNENRKVYKARQVKNYREMVEYSCKNYAQNIAYKYKKDYTAKNVEYIEKTYEQVG
Step 2 – Parameters
- Matrix: This option allows you to choose the scoring matrix to be applied to the search.
Default value is: BLOSUM62
Tip: In general, higher value BLOSUM matrices (e.g. BLOSUM90) and lower value PAM matrices (e.g. PAM30) are more stringent than low value BLOSUM or high value PAM matrices. This implies that if you want to find more distantly related homologues, you should preferentially employ a low value BLOSUM or high value PAM matrix (For more information about scoring matrices see http://en.wikipedia.org/wiki/Matrix).
Step 3 – Run
- Click "Submit" (or click "Reset" to clear your input).
Step 4 – Link BLAST output to BGCs
- Show BGC for select hit: Click the hit protein id to the BGC which contain this ID.
- Show BGCs in all hits: Click this button on the top of BLAST output page to get all matched BGCs.
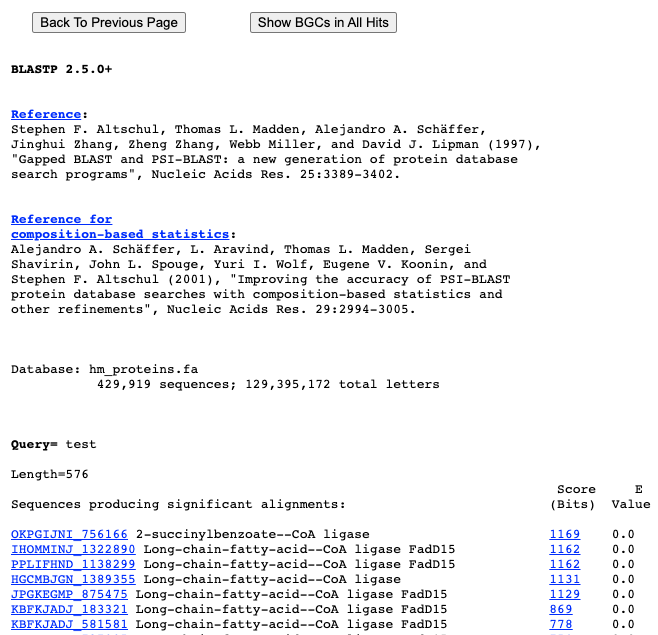
Browse
Our browse page serves as a useful tool for users to easily navigate and explore the diverse range of secondary metabolite BGC data available. With three options to choose from - browsing by Type, Phylum, or both - users have the ability to narrow down their search and discover specific data sets. However, it's important to note that the options of browsing by Type or Phylum are mutually exclusive, and can only be selected if the other option is set to 'any'. On the other hand, selecting both options will provide results that encompass both criteria. We hope this feature enhances your experience and helps you find the information you need.
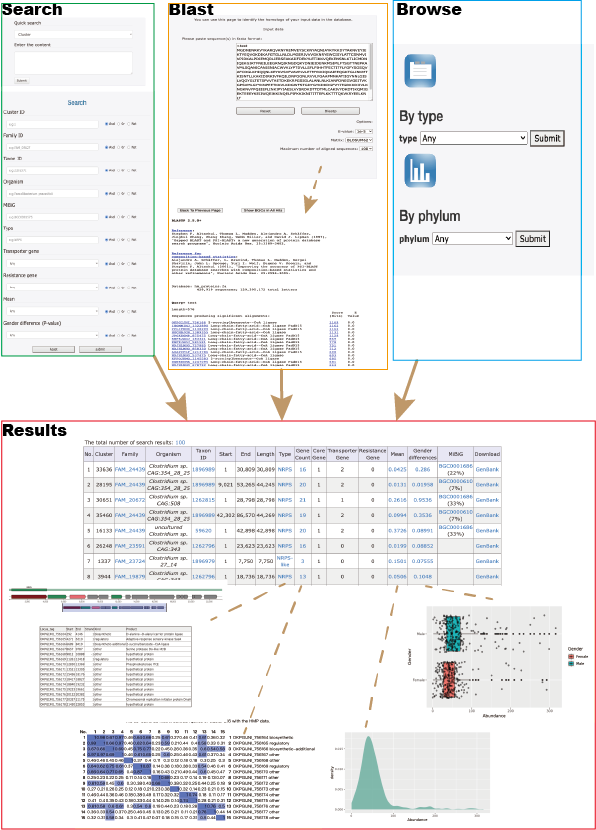
Allow us to provide you with an example to better understand the layout and functionality of our table. The table you see before you is the result of a search, blast, or browse action, and presents a clear and concise description of the data sets available. Each row represents a unique data set, and the various columns provide important information such as the CLuster, Family, and other relevant details. We hope this helps you make informed decisions and find the data you need efficiently.You can download search results as spreadsheets by clicking the total number of search results. Currently, the maximum number of results you can download is 2000.
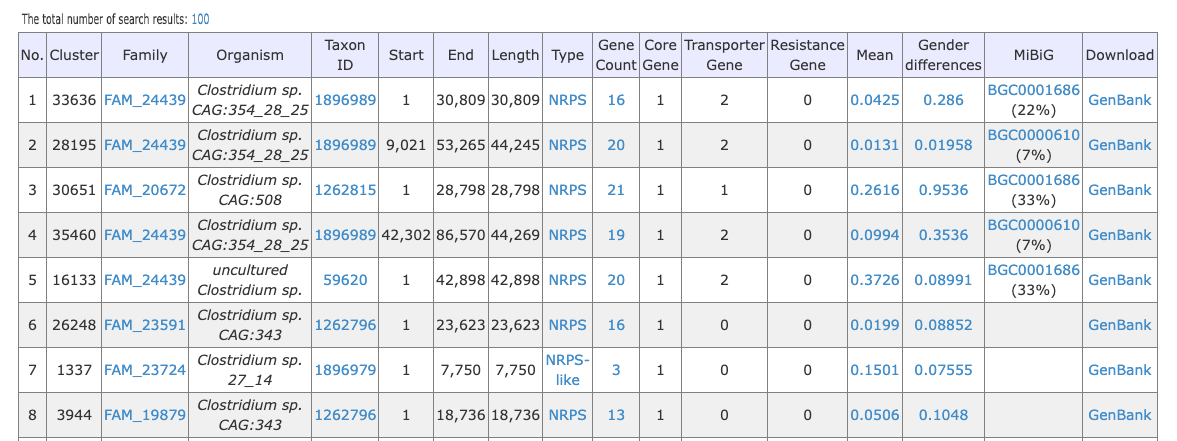
sBGC-hm provides the following information for a BGC.